The Challenge
The functions of genes are intensively studied from both a basic science and applied translational perspective. It is amazing that gene promoter variants that control expression of genes are not used in the clinical management of patients with cancer. Consider that there are over 70,000 scientific papers investigating promoters of cancer genes, dysregulation of tumor suppressor and protooncogene expression are established cancer mechanisms, and dozens of transcription factors controlling gene expression such as p53 and c-Myc are major cancer drivers.
Despite such a pervasive impact, the analysis of genetic variants in gene promoters is not yet used, but readily applied to the diagnosis and treatment of cancer. The key limitation is lack of extensive knowledge about promoter variants.
GMLs, the solution for determining the impact of VUS on function
Heligenics offers a new product with measured impact of single nucleotide variants in the promoter region of 60 key cancer driver genes. The licensees of this library will increase the accuracy and diagnostic yield of a genetic cancer test. Through a massively parallel in vivo process called the GigaAssay, Heligenics produces what we call a Gene Mutation/Function Library (“GMLs”). Each GML measures the impact of each nucleotide substitutions in the promoters on expression of many key cancer genes. To individually test each nucleotide substitution in each gene by other approaches in human cells would be impossible.
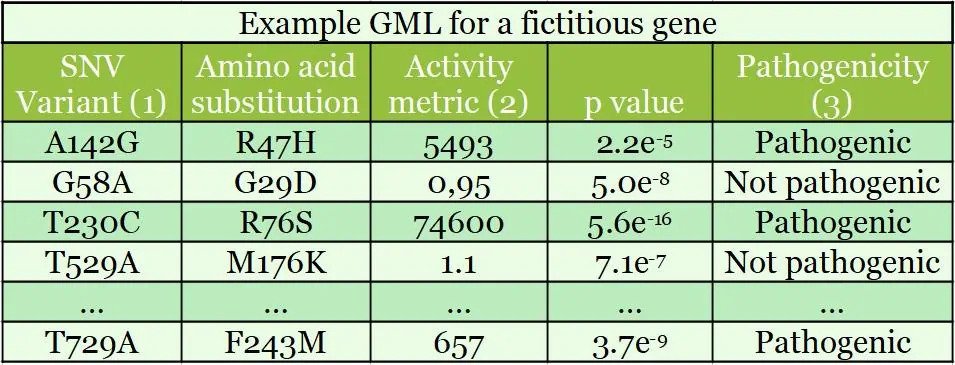
Shown above is a GML result for a fictitious gene; this table shows 5 variants from a table of over 10,000 SNVs (1). This type of GML has >97% of single nucleotide variants encoding single amino acid substitutions. For the Activity Metric (2), activity is measured against wild-type activity in the first column. A measurement of 100 is 1% of the wild-type (‘baseline’) activity. Pathogenicity (3) is determined from an activity threshold relative to wild-type activity and known pathogenic mutations.
Heligenics Gene Promoter Product Sheet Download
Kindly fill out the form below to access a handy PDF.
