The Challenge
All pharmaceutical companies have many drugs that have failed in a previous clinical trial or were not approved by the FDA. Anywhere from $800 Million to $1.4 Billion and many years have been spent on each compound. What if a percentage of these drugs or their derivatives could be brought to market? Imagine the benefit to patients with novel therapies that previously failed due to underlying genetic variants rendering them susceptible to adverse events. Unlock value from the previous investments straight to the company bottom line.
GMLs, the solution for drug revitalization
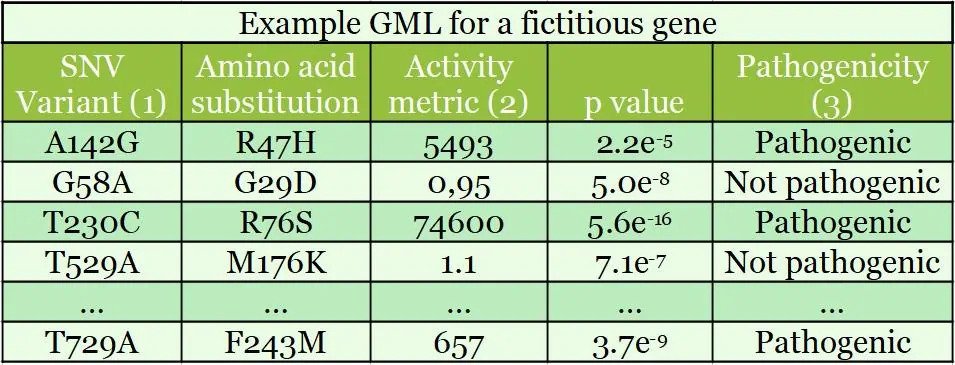
Heligenics offers a new product to revitalize these failed drugs by stratifying genetically defined populations to enhance efficacy and safety, thus minimizing the main failure points for clinical trials. Through a massively parallel experimental test of drug’s effect upon cells, Heligenics uses the “GigaAssay” to produces Gene Mutation / Function Library (“GMLs”). Each GML experiment measures the impact of all possible amino acids substitutions in the drug target upon the targets molecular function. This experiment is repeated in the presence and absence of the drug.
To improve efficacy and safety, comparison of GMLs identifies substitutions that can produce resistance, including those in the drug binding site, and those causing cell toxicity. The variants in the GML’s are next compared to population
Heligenics offers a new product to revitalize these failed drugs by stratifying genetically defined populations to enhance efficacy and safety, thus minimizing the main failure points for clinical trials. Through a massively parallel experimental test of drug’s effect upon cells, Heligenics uses the “GigaAssay” to produces Gene Mutation / Function Library (“GMLs”). Each GML experiment measures the impact of all possible amino acids substitutions in the drug target upon the targets molecular function. This experiment is repeated in the presence and absence of the drug.
To improve efficacy and safety, comparison of GMLs identifies substitutions that can produce resistance, including those in the drug binding site, and those causing cell toxicity. The variants in the GML’s are next compared to population
Technical approach
Many recently approved drugs (>140) have a genotype on the label, thus genetics is becoming increasingly important in drug testing. Heligenics GMLs will classify molecular function and cell toxicity of each variant, thereby inferring efficacy and safety for patients having any genetic variant in the drug target gene. Combined with allele frequency data and drug binding site data, clinical trials can be accurately designed.
Brivanib has had a massive investment and was tested for treatment of patients with hepatocellular cancer in 25 different clinical trials and has failed four Phase 3 trials, the last in 2017 (clinicaltrials.gov). In general the drug is well
tolerated, but is failing trials for its poor efficacy, especially when compared to Sorafenib, the only approved drug on the market. The patient response the drug is variable with some participants having a positive response and others with no effect.
The drug targets are 7 receptors in the tyrosine kinase family, three Vascular Endothelial growth Factor Receptors 1-3 (VEGFR), and four Fibroblast Growth factor Receptors 1-4 (FEFR). One target, VEGFR2, is illustrated below, and is the target for Brvanib.
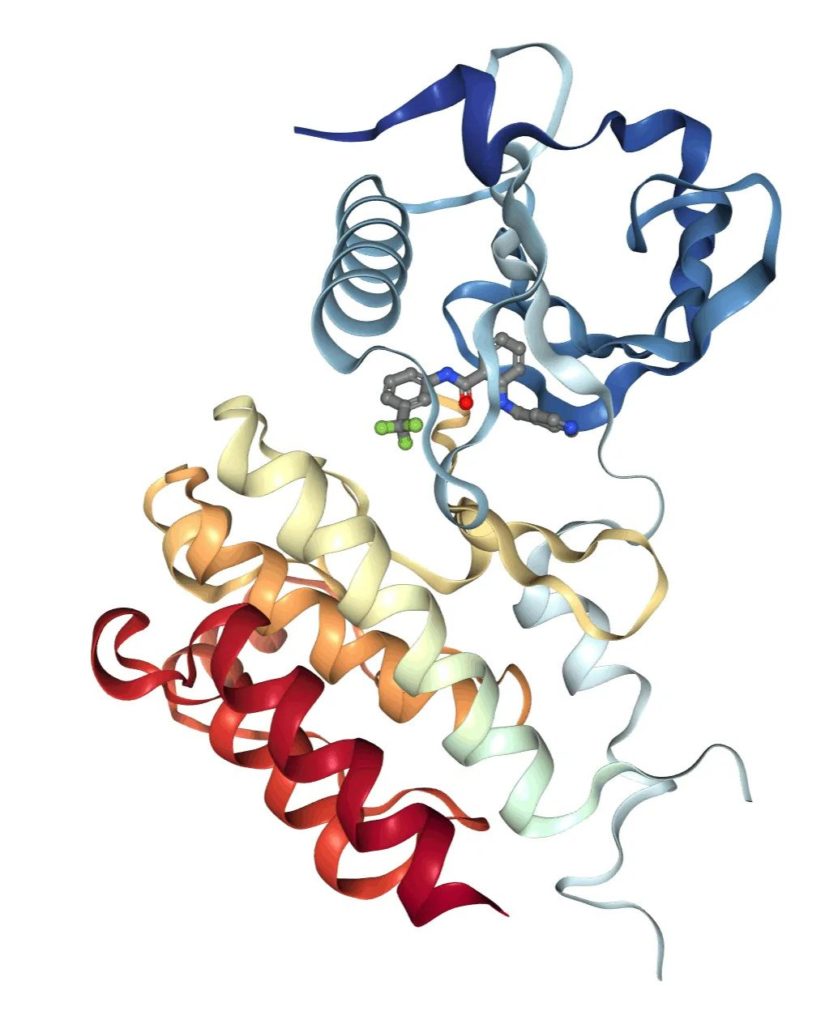
Protein Databank structure of VEGFR2 bound to AAL993 (PDB: 5EW3)
The structure for Brivanib complexed with a receptor was not available but there is a structure with another ATP analog, AAL993 that binds VEGFR2 (PDB: 5EW3). The contact residues for the drug with the binding site are: V848, A866, V867, K868, I892, E885, I888, L889, V898, V914, V916, E917, C919, L1019, L1035, I1044, D1046, F1047.
There are no mutations in VEGFR2 for hepatocellular cancer and only 17 variants for other disorders in ClinVar, the main public disease-variant database. However, there are 713 missense variants observed in ~250,000 people ranging from singletons to 22% allele frequency; 74 variants are in the drug binding site region. There are about 1,500 missense variants observed in cancers (COSMIC), with approximately 100 in this binding site region.